Researchers at Université de Montréal (UdeM) have made a significant discovery regarding the evolution of molecular systems crucial to the development of life. Through the linking of molecules, they have uncovered insights into the emergence of intricate self-regulating mechanisms.
Alexis Vallée-Bélisle, a professor at UdeM and the lead researcher of the investigation, explained that life’s sustenance on our planet stems from myriad nanostructures. These nanomachines have undergone evolution over countless years.
These nanostructures, frequently smaller than 10,000 times the width of a human hair, predominantly consist of proteins or nucleic acids. Some nanostructures are made from a single component, like linear polymers that fold into specific shapes. However, most are built from multiple components that come together on their own to form large and adaptable structures.
Dynamic Molecular Structures: Reactivity to Environmental Triggers
These molecular structures exhibit remarkable dynamism. For instance, they accurately react to diverse triggers such as fluctuations in temperature, oxygen concentrations, or nutrient availability, which leads them to activate or deactivate as required.
Just as cars need a series of steps—ignition, brake release, gear change, and gas input—to move forward, molecular systems also rely on a sequence of activation or deactivation of different nanomachines to carry out specific tasks. This includes everything, from basic movements to complex cognitive functions like thinking, explained Prof. Vallée-Bélisle.
These researchers posed a fundamental inquiry: how were dynamic molecular structures formed, organized, and optimized to sustain life?
Their discovery suggests that numerous biological formations may have arisen through the random linking of interacting molecules (such as proteins or nucleic acids like DNA or RNA) with linkers serving as a “connector” between each component.
Because biomolecular assemblies are vital for organisms to react to their surroundings, the scientists theorized that the way components are connected might also influence how their responses evolve dynamically over time.
Exploring Connectivity in DNA-Interacting Molecules
To investigate this, Dominic Lauzon, who was a doctoral student during the study, chose to synthesize and link numerous DNA-interacting molecules together. This allowed him to examine how connectivity affects the dynamic behavior of the assembly.
According to Lauzon, the highly adaptable and accessible chemistry of nucleic acids like DNA makes them ideal for studying fundamental questions about biomolecule evolution. Additionally, nucleic acids are believed to be the molecule responsible for the emergence of life on Earth.
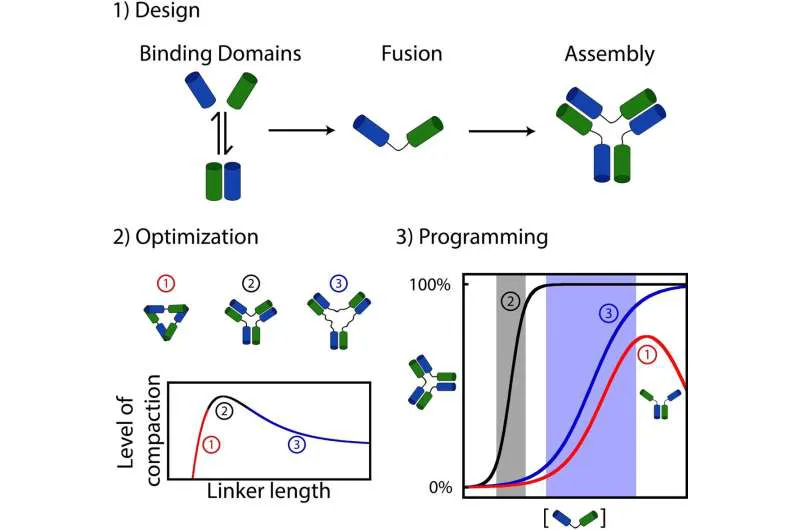
Linkers: Regulators of Assembly Dynamics
Lauzon and Vallée-Bélisle found that altering the length of the “linker” between interacting molecules resulted in notable changes in assembly dynamics. Some assemblies showed high sensitivity to stimuli variations, while others needed larger changes to promote assembly.
“Linker”, here refers to a chemical structure that connects two or more other molecules together, and facilitating their interaction or assembly.
Remarkably, certain linkers introduced new regulatory functions like self-inhibition, where a stimulus promoted both assembly and disassembly. These diverse responsive behaviors are reminiscent of those observed in natural living nanomachines.
Deciphering Assembly Dynamics
With the help of experiments and mathematical equations, the team was able to decipher the reason why a straightforward adjustment in linker length was highly effective in altering the dynamics of molecular assembly.
The molecules responsible for forming the most stable assemblies also induced the most sensitive activation mechanisms. Conversely, linkers associated with less stable assemblies led to less sensitive activation mechanisms, sometimes even causing self-inhibition, explained Lauzon.
Blueprint for Adaptable Nanosystems
Precisely detecting molecular signals is vital for both biological systems and the advancement of nanotechnology, which relies on recognizing and incorporating molecular data.
Consequently, the researchers suggest that their findings could offer a foundational blueprint for designing more adaptable nanomachines or nanosystems with finely controlled functions. This could be achieved, for instance, by attaching interacting molecules using different linkers. Such molecular assemblies are already being utilized in areas such as biosensing and drug delivery.
In addition to offering a straightforward approach to develop advanced self-regulating nanosystems, the researchers’ findings illuminate the potential origins of natural biomolecular assemblies and their optimal dynamics.
Takeaway
The development of self-regulating nanosystems based on the principles uncovered in this research could revolutionize various fields, such as medicine, electronics, and environmental monitoring.
Furthermore, this research provides valuable insights into evolutionary processes, offering a deeper understanding of how natural biomolecular assemblies develop their optimal dynamics.
Overall, the research has the potential to drive innovation across multiple disciplines and pave the way for new technologies and applications with far-reaching implications for society.