Researchers at Southwest University in China have successfully constructed the chromosomal-scale genome assembly as well as the complete spidroin gene set of the golden orb-weaving spider.
The chromosomal-scale genome assembly represents genome sequence; while the spidroin refers to proteins in spider silk. Different spidroins determine the varieties in spider silks.
With this new insight, researchers can dive deeper into the silk-producing capabilities of spiders. Eventually, this might lead to the development of new materials with desirable mechanical properties.
Other equally interesting offshoot of this research could be:
- in the understanding of evolutionary history of spiders,
- know-how into their silk production, and
- assistance in the conservation efforts for endangered spider species.
Remarkable properties of spider silk
Spider dragline silk has remarkable properties which makes it an attractive material for various applications, including medical and industrial.
Therefore, researchers think molecular understanding of the silk can lead to innovative approaches for producing spider silk proteins that are more efficient and cost-effective than traditional methods.
For instance, researchers are exploring the use of bacteria, yeast, and plants to produce spider silk proteins in large quantities, which could potentially overcome the scaling challenge.
Additionally, advances in genetic engineering techniques such as CRISPR-Cas9 could enable the creation of genetically modified organisms that produce silk proteins with improved properties.
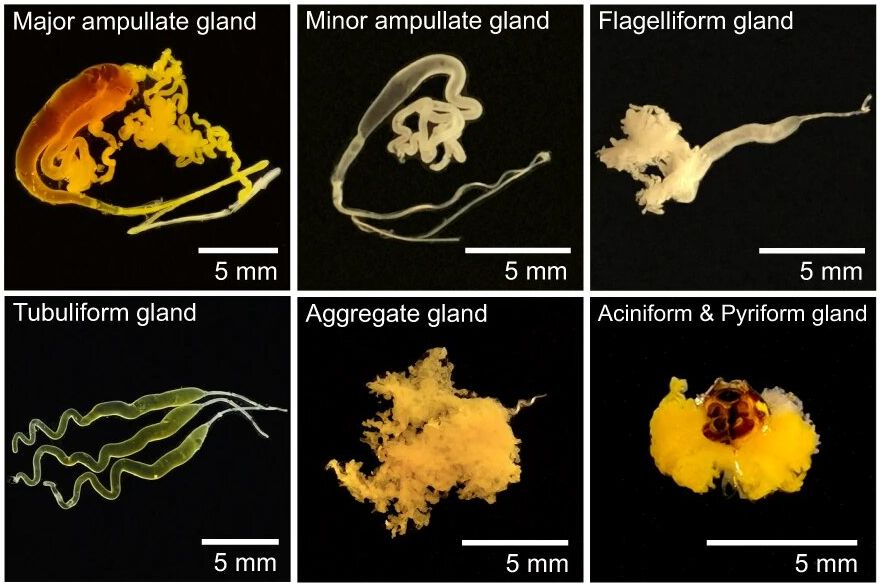
Multiple sequencing techniques to get into the genetic basis
To obtain the genome, researchers made use of multiple sequencing techniques.
Initially, the Oxford Nanopore platform was used. The platform is a next-generation sequencing technology that uses nanopore-based sequencing to produce long, contiguous reads of DNA or RNA molecules.
Followed by Illumina sequencing machines. They are a type of next-generation sequencing (NGS) technology that use a massively parallel sequencing approach to rapidly generate large amounts of DNA or RNA sequence data.
As well as the Hi-C for chromosome mapping. It is a method used to investigate the three-dimensional organization of chromosomes in the nucleus of a cell. It works by cross-linking chromatin segments that are in close proximity in the nucleus, then digesting the chromatin with a restriction enzyme, and ligating the cross-linked fragments together.
By combining these multiple sequencing techniques, the research team was able to generate a more comprehensive and accurate genomic dataset.
Analysing the different segments of the major ampullate gland also provides insight into the various stages of silk production, from gene expression to protein synthesis and secretion.
This information could help researchers identify key regulatory genes and proteins involved in silk production and potentially develop strategies for enhancing silk production or engineering silk proteins with specific properties.
Takeaway
The multidimensional approach taken by the researchers provides a more complete understanding of spider silk production and could lead to exciting new applications in the future.
Overall, the new molecular atlas of spider silk production is an exciting development that could pave the way for practical applications of this remarkable material in the future.
Via: Phys.org