Researchers at the University of Cologne (UoC) have achieved a groundbreaking milestone by creating artificial nucleotides. Nucleotides forms the building blocks of DNA. So, if we go by the research, the innovative development will make way for potential advancements in genetic engineering as well as molecular biology.
Before going further, let’s talk a li’l about the cell and the environment it holds. A cell contains some raw ingredients that is floating inside its cellular matrix. Within this soup of molecules, there also happens to be the base materials which is needed to make DNA and RNA. It’s like the early stages of Earth’s formation, all these “primordial elements” were sort of cooking up naturally in the mix.
Similarly, cells also figured out how to fabricate the DNA and RNA from the basic building blocks that were present in its microenvironment.
What is DNA?
DNA, shortened for deoxyribonucleic acid, is a molecule that contains the genetic information. This data forms the fundamentals for development, functioning, growth, and reproduction.
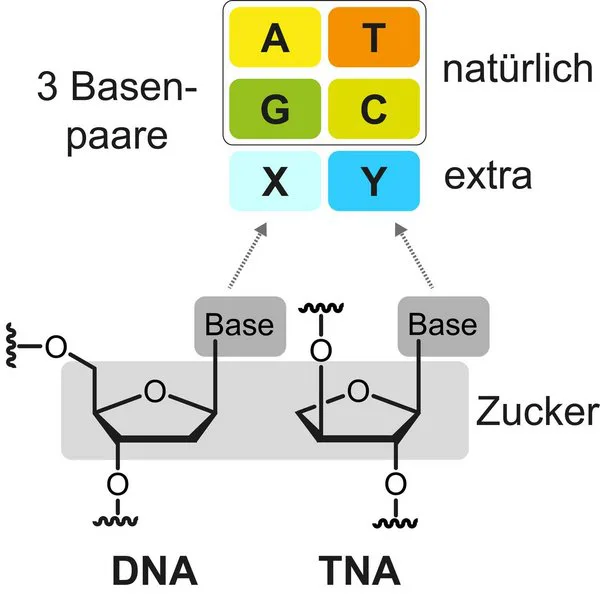
Basically, DNA consists of only four different building blocks, the nucleotides. The double helical structure is the polymer made up of repeated units of this nucleotide.
If we go at further, we will find, four distinct parts of a single nucleotide, which is:
- a sugar molecule (deoxyribose),
- a phosphate group, and
- one of four nitrogenous bases: adenine (A), thymine (T), cytosine (C), and guanine (G).
The sequence of these bases along the DNA strand carries the genetic information.
Synthetic Nucleic Acids
Researchers developed a novel nucleic acid called threofuranosyl nucleic acid (TNA). This molecule features an extra base pair.
In-depth information is provided in the research paper titled ‘Expanding the Horizon of the Xeno Nucleic Acid Space: Threose Nucleic Acids with Increased Information Storage’. It was published in the Journal of the American Chemical Society.
Creating Enzyme-Resistant and Replication-Competent Variants
Xeno nucleic acids (XNAs) represent synthetic variants of DNA or RNA. Predominantly, it features alterations in its structural composition. Conventionally, these modifications occur in singular segments of the molecule. However, the current research is about making XNAs with changes in multiple parts of their structure.
This time, the researchers have combined two types of modifications:
- First, they have made XNAs resistant to enzymes. Else the chemicals would easily break them down (called TNA).
- In second, researches have enabled the XNAs to replicate efficiently and accurately (using a pair of unnatural bases called TPT3:NaM).
The team was able to create new modified building blocks for XNAs with both modifications.
Subsequently, they conducted experiments to make sure that these modified building blocks were compatible with specialized enzymes for synthesizing XNAs.
Results revealed the successful construction of one variant of modified XNA, namely TPT3-modified TNA. However, the synthesis of another type, NaM-modified TNA, proved to be more challenging.
Takeaway
This exciting research represents the inaugural achievement in creating a brand new type of XNA called exTNA, which comes with an expanded genetic alphabet.
This innovation holds tremendous potential for developing innovative nucleic acid-based therapies. Especially in cases where nucleic acids are used to replace or modify defective genes in order to treat genetic disorders. Along with aptamer-based therapies.
The aptamer-based therapies harness the unique properties of aptamers, which are short nucleic acid sequences that can bind to specific target molecules with high affinity and specificity. It offers wide platform for developing highly specific and effective treatments for cancer, infectious diseases, and autoimmune disorders.