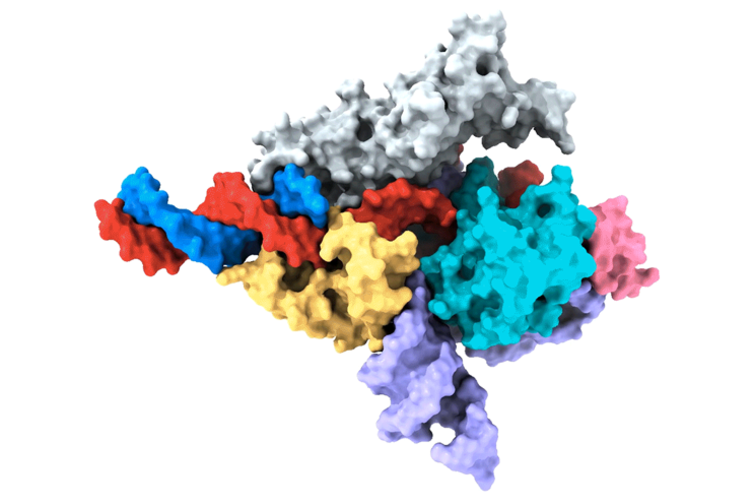
An international team of researchers has identified a programmable RNA-guided system in eukaryotes. Eukaryotes encompass plants, animals, fungi, and protists.
The newly discovered system is based on a protein called Fanzor. It uses RNA as a guide to precisely target DNA.
The researchers demonstrated that Fanzor proteins can be reprogrammed to edit the genome of human cells.
Compared to CRISPR/Cas systems, the Fanzor system is more compact and has the potential to be delivered more easily to cells and tissues as therapeutics.
RNA-guided DNA-cutting mechanisms across all kingdoms of life
CRISPR/Cas systems were initially discovered in prokaryotes. And researchers were curious to know if similar systems existed in eukaryotes as well.
The study’s senior author, Feng Zhang, stated that the Fanzor system provides an additional way to make precise changes in human cells. Thus, complementing the existing genome editing tools, that is CRISPR/Cas systems.
The research, therefore, confirms the presence of RNA-guided DNA-cutting mechanisms across all kingdoms of life.
Exploration beyond the CRISPR system
The Zhang lab, from where the study has originated, has a primary goal of developing genetic medicines by utilizing systems that can modulate human cells through targeted gene manipulation.
Beyond the widely known CRISPR system, the lab has been exploring other RNA-programmable systems present in nature.
Nearly two years ago, the lab was able to bring about an important discovery. The team identified a class of RNA-programmable systems called OMEGAs in prokaryotes (bacteria and other single-cell organisms lacking nuclei).
These OMEGA systems were associated with transposable elements or “jumping genes” found in bacterial genomes. They are believed to have given rise to the CRISPR/Cas systems.
Not only this, the research also highlighted parallels between the prokaryotic OMEGA systems and the Fanzor proteins identified in eukaryotes. Thus, suggesting that Fanzor enzymes might employ an RNA-guided mechanism to target and cleave DNA.
The finding concluded that similar RNA-guided DNA-cutting mechanisms are present in both prokaryotes and eukaryotes.
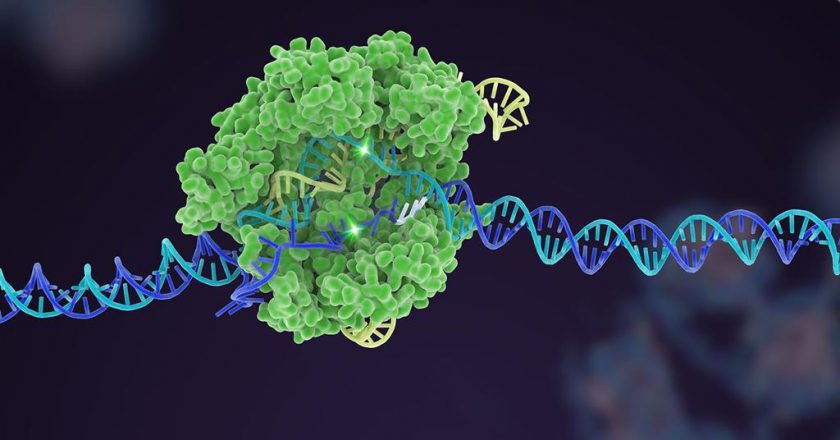
Fanzor proteins utilize ωRNAs for precise DNA targeting
A new study was conducted, this time, it was on RNA-guided systems. It focused on Fanzors found in various organisms including fungi, algae, amoeba species, and the Northern Quahog clam.
Makoto Saito, from the Zhang lab, led the biochemical analysis of Fanzor proteins.
He observed that these proteins are enzymes called DNA-cutting endonucleases. They utilize specific non-coding RNAs called ωRNAs to target specific locations in the genome.
This mechanism, observed in animals, is the first of its kind discovered in eukaryotes.
In contrast to CRISPR proteins, Fanzor enzymes are encoded within transposable elements in the eukaryotic genome.
The team’s phylogenetic analysis suggests that Fanzor genes have transferred horizontally from bacteria to eukaryotes.

Takeaway
The exploration of these RNA-programmable systems beyond CRISPR/Cas showcases the Zhang lab’s efforts in uncovering the diverse range of genetic tools found in nature.
A programmable RNA-guided system in eukaryotes expands the possibilities for genome editing and offering potential advantages over existing CRISPR/Cas systems.
Such discovering not only provide the potential of RNA-guided systems for precise gene editing in various organisms. But they are also insightful for the development of advanced genetic therapies.